-
- Downloads
some small fixes and tests for BioTree.
Showing
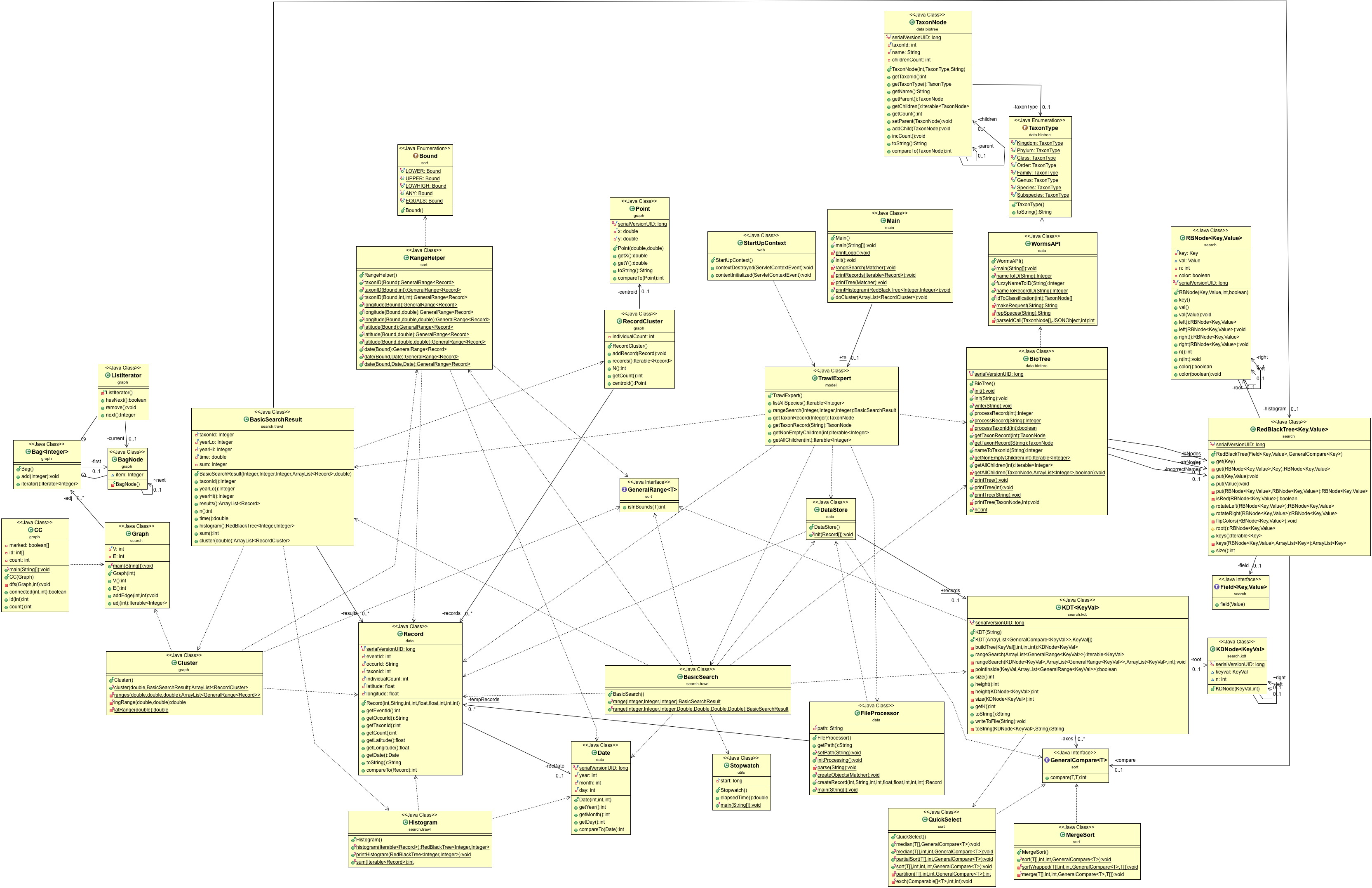
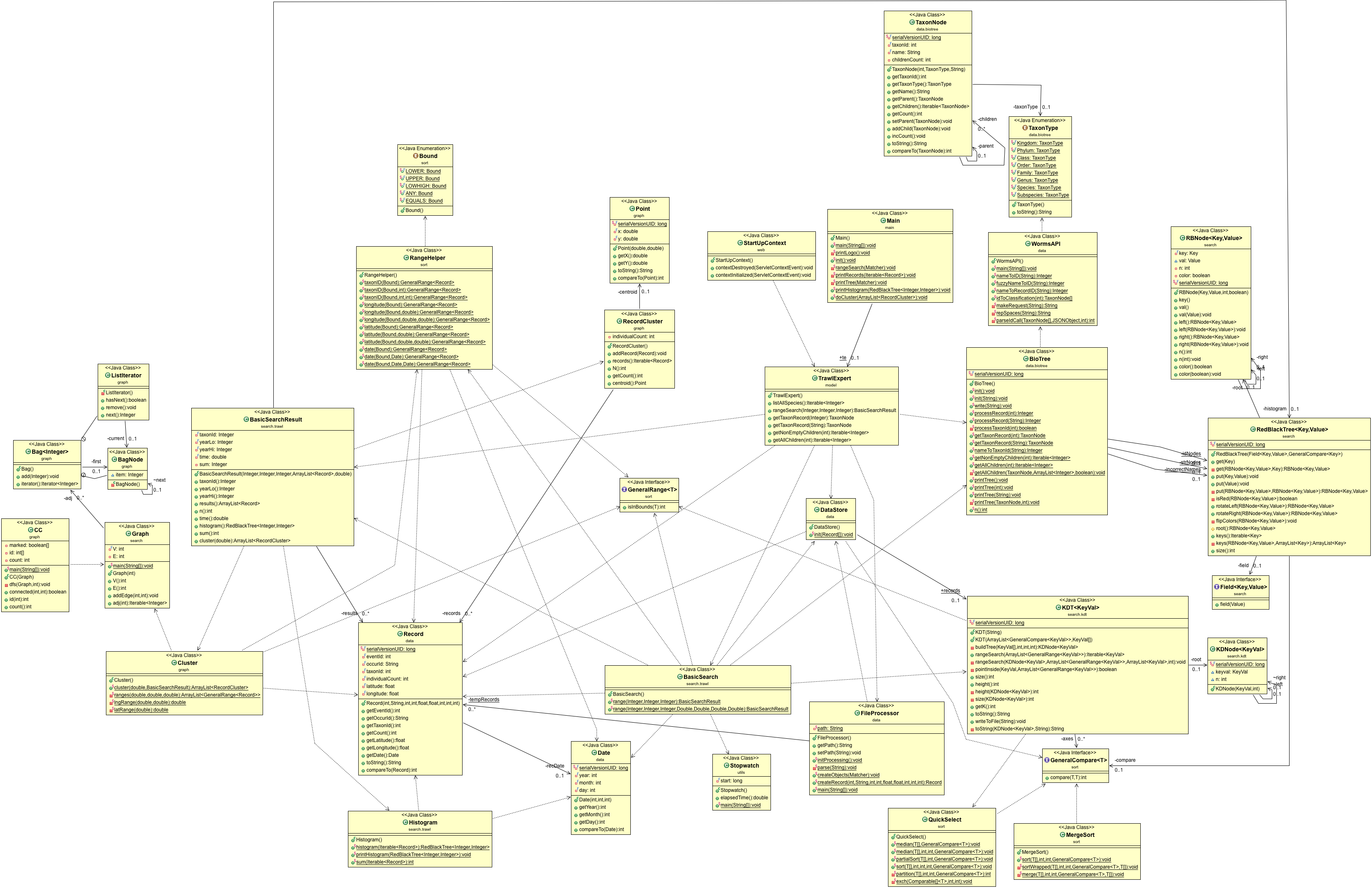
- UML.png 0 additions, 0 deletionsUML.png
- designSpec/designSpec.pdf 0 additions, 0 deletionsdesignSpec/designSpec.pdf
- src/data/FileProcessor.java 1 addition, 1 deletionsrc/data/FileProcessor.java
- src/data/TestBioTree.java 0 additions, 39 deletionssrc/data/TestBioTree.java
- src/data/biotree/BioTree.java 9 additions, 3 deletionssrc/data/biotree/BioTree.java
- src/data/biotree/TestBio.java 76 additions, 0 deletionssrc/data/biotree/TestBio.java
- src/data/biotree/TestBioTree.java 49 additions, 0 deletionssrc/data/biotree/TestBioTree.java
- src/model/TrawlExpert.java 4 additions, 2 deletionssrc/model/TrawlExpert.java
| W: | H:
| W: | H:


No preview for this file type
src/data/TestBioTree.java
deleted
100644 → 0
src/data/biotree/TestBio.java
0 → 100644
src/data/biotree/TestBioTree.java
0 → 100644